The Shigatoxigenic Escherichia coli (STEC) are bacterial enteropathogens responsible for some intensive clinical syndromes such as bloody diarrhoea, hemorrhagic colitis, hemolytic uremic syndrome, thrombotic thrombocytopenic purpura, and renal failure. These pathotypes come under the Enterohemorrhagic Escherichia coli (EHEC) group. Monogastric farm animals such as pigs, horses, chickens, ducks, turkeys and aquatic animals like shellfish, fishes, and wild animals can act as major spillover hosts of STEC strains and could serve as the potential source of infection. The pathogen is notorious as a quickly emergent strain with acquired characteristics like different variants of Shigatoxin, many antibiotic degrading enzymes, Intimin, Enterohemolysin, Auto-agglutination Adhesins, Catalase-peroxidase, Zinc metalloprotease, Subtilase cytotoxin, tolerance to multiple adverse conditions, and biofilm formation. The bacteria are known for its long survival in different adverse physical-chemical conditions. The formation of biofilm is one of the major factors responsible for their persistence. Multidrug resistance is another related trait contributing to the high mortality rate of these strains. STEC strains are good candidates for studying the emergence of pathogens with acquired characteristics like genes. In this article, various virulent traits and multidrug resistance that enabled the strain to emerge as a serious public health menace were reviewed.
STEC, Antibiotic Resistance, Shiga Toxin, Review, Horizontal Gene Transfer
Some pathotypes of Escherichia coli have gained attention as a significant foodborne pathogen by acquiring morbid genes from similar and related bacteria through horizontal gene transfer mechanisms. Regarding the pathogenicity, the Escherichia coli are categorized into six classes, namely Enterohemorrhagic type (EHEC), Enterotoxigenic type (ETEC), Enteropathogenic type (EPEC), Enteroaggregative type (EAEC), Enteroinvasive type (EIEC) and diffusely adherent type (DAEC).1 Out of these categories, the enterohemorrhagic one is a clonal class with somatic O antigen called Shigatoxigenic Escherichia coli or Verocytotoxin-producing Escherichia coli (STEC or VCEC). Shigatoxigenic E. coli., with cattle as principal natural reservoirs, especially ruminating post-weaning calves and heifers, is an essential group of bacterial enteropathogens. E. coli O157:H7 is highly popular as well as familiar to both bacteriologists engaged in diagnostic field and the common public.2 These STEC strains are highly responsible for some severe clinical presentations, chiefly bloody diarrhoea, hemolytic uremic syndrome, hemorrhagic colitis, thrombotic thrombocytopenia purpura, and renal failure.3-5
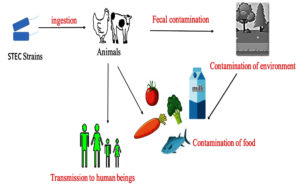
The pathogens cause asymptomatic infection in cattle. In cattle, the vascular receptors are absent in the intestinal mucosa, and thus the endocytosis and transportation to other organs are inhibited, resulting to the asymptomatic colonization of STEC strains in the large intestine.6-8 In addition to cattle, other ruminants like sheep and goats are also identified as remarkable carriers and asymptomatic shedders of O157 and different strains in the epidemiological studies conducted in the United States, Australia, as well as Europe.9,10 Various studies conducted have provided strong proof for their occurrence in farm animals such as pigs, horses, chickens, ducks, and turkeys which could act as a significant spillover host of STEC strains and can be the potential source of infection.11,12 Many diverse studies have been conducted to bring out the pervasiveness of STEC among various species and identified that rodents such as rats as well as pigs, many wild bird species, fish, shellfish and insects such as house flies could be relevant vectors for the dissemination of STEC infection.13 The STEC outbreaks associated with the intake of fecal-contaminated food are on the rise.14,15 Contaminated foods of bovine origin, milk and plant-based foods are often implicated in foodborne outbreaks (Figure 1).16
Figure 1. Transmission of Shigatoxigenic E. coli.16 STEC transmitted to human beings through various sources such as animals and fecally contaminated food and water
The outbreaks by these isolates are typically seen as associated with contaminated beef, i.e. in the food industry, especially in food-processing plants, STEC constitute a significant concern and are mainly associated with the contamination of beef carcasses. The contamination may occur during different processing stages, such as slaughtering, dressing, and chilling.17 These bacterial populations are more likely to be present on the surface of the equipment involved in processing. The potentiality to develop biofilm on both living and non-living surfaces is considered as the primary factor responsible for the persistence of STEC strains in meat processing plants. Studies found that biofilms exhibit higher resistance to disinfectants such as quaternary ammonium compounds.18 In a food industry environment, biofilm development greatly benefits the microbial cell population in many ways. It provides physical resistance against desiccation, chemical protection against various antimicrobials and disinfectants, as well as mechanical resistance against the liquid streams in pipelines used in the industry.19 The bacterial species involved in biofilm formation can undergo genomic variations with respect to crucial genes involved in biofilm formation, resulting in the construction of entirely different biofilms under altered conditions, thus complicating the eradication process in the food industry.
Moreover, the biofilm-forming ability of a bacterial strain is related to numerous factors. The bacterial cells can reach the desired surfaces mainly by flagella-assisted or twitching motility and indirectly promote biofilm formation.20 The adherence factors, such as curli and fimbriae eaeA, help establish initial colonization by facilitating the attachment on the target surface, followed by the formation of exopolysaccharide matrixes and finally, the three-dimensional structure of the biofilm is formed.21 The final structure of the biofilm is assisted by the cell to cell communication, occurring through quorum-sensing molecules.22 Thus, biofilm significantly contributes to the virulence mechanism of the strains. In addition, many other virulence factors have also been involved. Though the precise mechanism of pathogenicity is not fully elucidated, shigatoxin and some adherence factors like Intimin encoded by eae gene are significant virulence factors.23,24
Evolution of antibiotic resistance in stec strains
Studies found that the stx2 genes are more prevalent in the genome of E. coli than stx1. Out of the variants identified the stx2a are most commonly seen followed by stx1a and stx2c (Table 1). The rest of the variants are having minor prevalence. Another characteristic feature noticed is that majority of strains are carrying a single stx gene. The stx genes are also seen in combinations as stx1a/stx2a, stx2a/stx2c and stx1a/stx2c.25
Table (1):
Distribution percentage of Shigatoxin gene variants25
No. |
Stx genes |
Percentage of distribution |
|---|---|---|
1 |
Stx1a |
35.4 |
2 |
Stx1c |
1.5 |
3 |
Stx1d |
0.8 |
4 |
Stx2a |
41.2 |
5 |
Stx2b |
1.9 |
6 |
Stx2c |
15.0 |
7 |
Stx2d |
2.7 |
8 |
Stx2e |
0.4 |
9 |
Stx2f |
0.8 |
10 |
Stx2g |
0.4 |
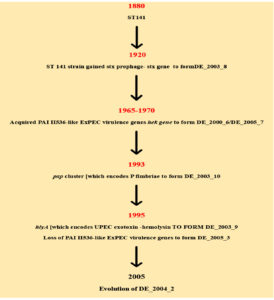
Several precise pieces of evidence exist for the constant evolution of antibiotic resistance in STEC strains (Figure 2). Studies showed that after reporting the initial STEC outbreak caused by O157strains in Japan, a steady rise in STEC infections was seen in the coming years.26,27 Moreover, studies revealed that the entire shigatoxigenic strains, non O157 strains as well as O157, were initially sensitive to the following antibacterial agents namely principen, trimethoprim, tetracycline, kanamycin, nalidixic acid, etc. were later found showing an increased frequency of resistance.28 Moreover, this rise in antibiotic resistance was observed not only in O157 strains but also in non-O157 strains, isolated mainly from the domestic animal reservoirs, which can, in turn, cause serious impact on other mainly food and environmental sources.28-30
Figure 2. Flow chart showing the evolution of STEC.26 The flow chart based on studies explains the process of evolution in shigatoxigenic E. coli. Initially, the insertion of prophage with stx gene in to the bacterial genome occurs. This is then followed by the addition of PAI II536 like EXPEC virulence gene, hek gene to form DE_2000_6/DE_2005_7 strains. Years later acquired the pap cluster which encodes P fimbriae to form DE_2003_10 strain. The hlyA gene encoding the UPEC exotoxin-hemolysin was acquired to form the strain DE_2003_9.This is followed by the partial or complete loss of PAI II536-like ExPEC virulence genes to form DE_2005_3 strain
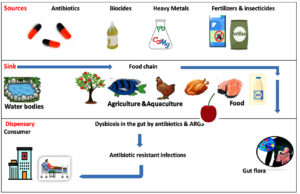
Antibiotics, biocides, heavy metals, insecticides, and fertilizers are selected for antibiotic-resistant genes and cause the contamination of animals, fish and plant contamination. These contaminated food items are consumed by humans and end up in their guts. Gut provides a very conducive environment for the spread antibiotic-resistant genes to the entire gut microbiota especially through horizontal gene transfer mechanisms. The endpoints of this antibiotic resistance transmission are the emergence of human pathogens, which leads to the development of antibiotic-resistant infections.32
Biofilm and multidrug resistance
Drug resistance and virulence factors are not only greatly important for the survival of the organism in the host body but also to overcome adverse environmental conditions. A diminished sensitivity exhibited by biofilms to antimicrobial agents is hypercritical trouble encountered in treating chronic infections. Moreover, the biofilm matrix provides a suitable environment for its residents to adjust to even higher concentrations of antimicrobial substances. The drug resistance mechanism exhibited by the biofilm population is highly distinguishable from planktonic forms and shows 1000 times more antibiotic resistance than planktonic forms.33
Besides the multicellular nature of biofilm, the partial or slow drug penetration and development of a micro-gradient in the concentration of the main metabolite may result in the formation of different phases, sluggish grower cell phase or stationary cell phase, adaptive stress response exhibited by some microbial cells, and differentiation of a little snippet of these cells to an extremely preserved antimicrobial resistant ‘persister-state’ are the four hypothesized resistance mechanisms of the biofilm community.34 The delivery of inactivated or sequestered antibiotics by reaction or binding to the interior of the biofilm can be decelerated completely. The interaction of the biofilm matrix polymers (negatively charged) with the aminoglycosides (positively charged) can also decrease the penetration of these substances.35 The transformed chemical microenvironment, together with the appearance of other microbial subpopulations in the biofilm matrix, is also accountable for the antibiotic resistance exhibited by the biofilm community.36
Another finding is that the bacterial population in a single species biofilm shows heterogeneity in their growth phases. This heterogeneous population include a gamut of swiftly expanding to a completely dormant type. If we consider a biofilm formed by a particular kind of organism, it is surprising that this population also shows heterogenicity in their growth states, as mentioned above. These metabolically inactive non-growers are capable of surviving the antimicrobial challenges.37,38
One important finding is that promulgation of the antimicrobial resistant mechanism crop up by the vertical transmission through DNA replication as well as horizontal gene transfer, mutation and inactivation or destruction of antibiotics through enzyme modification. In STEC strains, antimicrobial resistance is disseminated mainly through horizontal gene transfer through conjugation, transduction and transformation. The multiple plasmids that belong to the major replicon types, such as IncF, and IncA/C, are found associated with this horizontal gene transfer. The overuse of antibiotics as food additives in animal husbandry can induce antibiotic resistance, which is subsequently transferred to enteric normal flora through conjugation, thereby creating a threat to both human and veterinary (Figure 3).31,39 The antibiotic resistance developed by these biofilm communities contribute to chronic infections imposing great challenge to the conventional antibiotics.37
Figure 3. Selective flow of ARGs. The dissemination of Antibiotic-Resistant Genes from the various environmental sources to Dispensary31
Many studies have proven that the biofilm is exhibiting a remarkable degree of resistance to antibiotics, disinfectants, antiseptics, etc. The main characteristic feature of biofilm is that an exopolysaccharide matrix can act as a strong adsorbent, barricade of diffusion, molecular strainer, etc. offering restricted penetration.37 Several studies show that competition for nutrients in mixed biofilms results in nutrient deficiency. It is found that this nutriment paucity again bestows the property of multi-drug resistance to these communities. Many antibiotic degrading enzymes are produced in adherent biofilms, which are captured and condensed in the polymeric matrix. For example, the enzyme Beta lactamases from E. coli is capable of hydrolyzing the Beta lactam ring in Penicillin and its derivatives, cephalosporin, as well as monobactams, thereby suggesting the requirement of alternative treatment measures.35
Another reason for this increased antibiotic resistance of the biofilm community may be the change of molecular targets of antibiotics, that is these targets are reprogrammed or modified thereby reducing the binding affinity of many antibiotics. The mechanism focuses on altering the natural, i.e. the original targets of antibiotic not altering the functions of protein through the mutation of DNA, methylation of ribosomal RNA which inhibits the proper binding of antibiotic molecules.40 One example is that the mutations occurring in gyrase and subunit B of RNA polymerase led to the development of resistance to fluoroquinolones and rifampicin, respectively. Some bacteria can use mutation-induced mechanisms facilitating the active transport of antibiotics to outwards as well as to the periplasm of the cell through the efflux-pumps. Here the mutation occurs either in the genes of chromosome or plasmids involved in the coding of these pumps.41
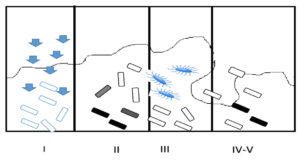
A unique organization in bacteria, named bacterial integrons incapable of self-transfer, showing multidrug resistance has been recognized and is hypothesized to perform a crucial function in procuring and propagating the drug resistance genes.42,43 They associate themselves with insertion sequences of transposons or conjugative plasmids for their transmission. These mobile DNA elements possess a specific structure with two conserved fragments adjoining a central region bearing cassettes, coding for drug resistance. Four categories of bacterial integrons have been recognized.44 Out of these, Class I integrons are more abundant. Their structure has two conserved fragments, namely 5’segment and the 3’segment. The 5’fragment can encode an integrase gene (intl1) to recruit genes for antimicrobial resistance. The 3’conserved segment bears qacEΔ1, encoding resistance to chemicals such as antiseptics and disinfectants.42,45 In addition to antibiotic resistance, biofilm bacteria also exhibit high resistance to chemical disinfectants. The reason behind this broad resistance is the presence of a small fraction of persisters, a subpopulation occurring in a hugely shielded condition, maybe similar to spores in the biofilm matrix. Persisters may be either triggered or spontaneous type constituting almost a very minute portion of the population. The frequency of this persister population is relatively high in the biofilm population compared to planktonic forms (Figure 4).45,46
Figure 4. Biofilm Resistance Mechanisms: (I) Slow or incomplete Antibiotic penetration; (II) Formation of concentration gradient of a metabolic substrate/product results to regions of slow or non-growers (shaded cells); (III) Expression of Adaptive stress responses (marked cells); (IV-V) Formation of persister state (dark cells)45
Main virulence factors
Shiga toxins
Although non-motile variants are occasionally isolated the O157:H7 is the major serotype responsible for the production of toxin. Most E. coli O157 isolates produce Shiga toxin 2; occasionally, Stx-1 as well as Stx-2 producing strains are seen, but Stx1 producers seen rare.47,48 Studies found that stx-1 has three subtypes listed as stx 1a, stx 1c and stx 1d. There are a total of seven subtypes of the stx-2 group, namely stx2a, stx2b, stx2c, stx2d, stx2e, stx2f and stx2g (Table 2).49,50
Table (2):
Main Virulence factors of STEC strains
No. |
Virulence Factors |
References |
|---|---|---|
i |
Shiga toxins
|
47 |
ii |
Surface factors
|
48 |
iii |
Virulence factors &mechanisms
|
49-51 |
After the initial adherence to the mucosal lining of the intestinal wall, the bacteria grow and start secreting several extracellular products, including a potent cytotoxin, Shiga toxins. Stx-2 has enormous number of variants, whilst Stx-1 is homogeneous. Even though, the two toxins are sharing about 60% of aminoacids and exhibiting DNA homology they show uniqueness in serological characteristics. These exotoxins, the most potent neurotoxin, are the virulence factors expressed by some bacteria such as Shigella sps. as well as several serotypes of E. coli51 (p201). These toxins share the same structure. Shiga toxins are type II ribosome-inactivating proteins.52
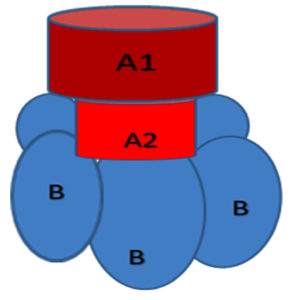
Shigatoxin comes under the protein toxin family namely, AB5 family. It has one A moiety showing the enzymatic activity and one non-toxic B subunit that enchains to cellular receptors. We can see five identical subunits of the B moiety form a pentameric ring that surrounds a central pore. The C-terminus of the A moiety is anchored in the central pore. Each B subunit possesses three specific binding sites. These binding sites interact with the trisaccharide component of the glycosphingolipid Gb3 specifically (Figure 5).52
Figure 5. Structure of AB5 Toxin. Shigatoxin belongs to the AB5 family of protein toxins with one A moiety that is enzymatically active and a B moiety with five identical subunits forming a pentameric ring surrounding a central pore that is nontoxic and responsible for binding to cellular receptors52
An endocytic process mediated by a receptor internalizes the exotoxin A receptor-mediated-endocytic mechanism internalizes the toxin molecule after binding to the target cell. In some cases, the toxin vesicles fuse with the lysosomes leading to their deterioration. But in some other cases, after the endoplasmic reticulum and Golgi complex processing, the subunit A is `nicked’ by an enzyme protease. This generates an A1 fragment of 27-kDa catalytically active and an A2 subunit of 4 -kDa. This A1 shows RNA N-glycosidase enzyme activity leading to the cleavage of a specified bond seen in the 28S rRNA, preventing the attachment of the aminoacyl-tRNA to the larger subunit of the ribosome, thereby blocking the process of elongation in protein synthesis resulting in programmed cell death.52 The Shigatoxins exert their toxicity by catalytically inactivating the 60S subunit of the ribosome. This can efficiently inhibit the translation in target cells by excluding the Adenine residue of the 28S rRNA.53,54 Shiga toxins can also traverse the boundary of epithelial cells and enter the blood vascular system, thereby reaching the kidney. This results in the destruction of glomerular endothelial cells expressing Gb3 leading to HUS development.55
Surface structures for biofilm formation
Biofilms possess a microbial multicellular lifestyle and are organized communities of bacteria interacting among themselves. Biofilm is attached to support either an inert or living surface contained in an extracellular polymeric substance.
Flagellum
The strains of E. coli possess peritrichous flagella for motility. In the investigations regarding the role of flagella in the formation of biofilm, it is found that the bacterial cells that lack flagella or with defective flagella, that is paralyzed type, face severe hindrance in biofilm formation The direct role of flagellum as an adhesin to help the cells to attach to the surface is not yet known. Several authors reported that H7 flagella and flagellin protein monomers are capable of binding to mucins and fresh bovine mucus. Another indication is that the removal of fliC genes made the bacteria less adherent than the wild-type parental strain. This finding strongly supports the adhesive property of H7 Flagella. The ability of H7 flagellum to act as an adhesin and its role in initiating colonization in cattle reservoirs is confirmed through several studies.56,57
Fimbriae
The Type-1 Fimbriae is the ubiquitous adhesins seen in Escherichia coli isolates, pathogens, commensals, and other Enterobacteriaceae. These are rod-shaped adhesive surface organelles with 7-nm width and a length of approximately 1 µm. It is seen to be associated with adhesion on host tissue. The structural components of fimbriae, fimbrial biosynthesis machinery and regulatory elements are encoded by fim gene cluster. In fim mutants, the analysis of biofilm formation showed defectiveness in the primary adhesion to many inanimate substrates like polyvinyl chloride in a rich culture medium under stagnant culture conditions. It is studied extensively and is considered as one of the best adhesins mediating the adherence of the bacterial cell to the surface of intestinal epithelial cell of the host.58 A 21-kDa protein is the main constituent of this fibril and was yag Z gene-encoded. These are designated as ECP (E. coli common pili) because of their wide presence in both commensal and pathogenic E. coli. The mutagenesis studies detected a putative fimbrial operon.56 Their expression in the E.coli strain, K-12 generated a long visible fimbriae of 1 to 2 µm long. It is seen prolonging from the bacterial cell and is capable of developing longer bundles dissimilar to flagella and was named F9. The studies detailed the involvement of F9 in the attachment to epithelial cells, fibronectin, etc. of the bovines.59
Autotransporter proteins
Another one is the autotransporters, a group of secretory proteins fulfilling all the demands necessary for the secretion across the outer membrane, cytoplasmic membrane as well as to the cell surface of the bacteria. Using tools such as autotransporter motifs researchers have found several autotransporters. The AIDA (Adhesin Involved in Diffuse Adherence), Tiba protein and Ag43 are some auto transporter proteins produced by different strains of E. coli. These auto-transporter proteins enhance biofilm formation and mediate auto aggregation.56,60 A self-recognizing surface adhesin, Antigen 43, can be seen in most strains. It exhibits an excellent character, that is, cell-to-cell aggregation, thus conferring clumping and fluffing of microbial cells. Thus, the key function of Ag43 is the promotion of biofilm formation on abiotic surfaces.61
Curli Formation
The Curli is a filamentous heteropolymeric proteinaceous appendages, composed of two monomers or subunits, major and minor, i.e. CsgA and CsgB, respectively. The attaching ability of majority of E. coli strains are influenced by curli. The major subunit, CsgA, produces Curlin and the minor subunit, CsgB, catalyzes the development of these curli on the surface of the bacterial cell, thereby acting as a surface-exposed nucleator.62 The expression of curli in most Enterotoxigenic and Enterohemorrhagic types, among clinical E. coli isolates. This suggests the specific role of curli in pathogenicity.63 The optimal expression of curli can be seen under low nutrients, the low osmolarity of the medium, temperature below 30°C and during the stationary growth phase of bacteria.64
Extracellular polysaccharide
Most E. coli strains produce an extracellular polysaccharide, M antigen or Colanic acid capsule. Under stress conditions, such as desiccation or osmotic upshifts, colanic acid protects bacterial cells. The wca cluster (cps) consists of 19 genes required for colanic acid synthesis65 The capsule consists of galactose, glucose, and glucuronic acid. The synthesis is controlled by Rcs system, the two-component regulator of capsule synthesis. The three core proteins of the system are Rcs C (a transmembrane sensor kinase), Rcs D (a transmembrane protein), and Rcs B (the response regulator).66
Bacterial conjugation
Bacterial conjugation includes a very close cell-to-cell interaction, where the transfer of a conjugative plasmid occurs between two bacterial cells with a specialized pilus seen on the donor.67 These plasmids have the complete set of genes necessary during both the horizontal and vertical transfer for their maintenance. The plasmid backbone has all the functions encoded in it. Studies on these conjugative plasmids has showed very much diversity in genetic properties as well as organization. This gives an indication that various regulations and molecular mechanisms as well as strategies are adopted by plasmids for horizontal gene transfer and maintenance. Studies also revealed that this F pilus can function as adhesion factor and allows imprecise contacts such as cell-stable surface connection or the cell-to-cell contact. This accelerates the initial adhesion to the abiotic surfaces, thereby substantiating the development of the 3D structure of the biofilm.68
Additional virulence markers
Intimin
The potentiality of producing the attaching and effacing lesions (A/E lesions) on several cell categories is one of the remarkable features of STEC strains. Intimin, outer membrane adherence protein, is a typical virulence factor essential in the attachment of bacteria to the epithelial cells. It is found that the Intimin interacts with Tir (Translocated intimin receptor) leads to the development of A/E lesions. These lesions play a crucial role in pathogenesis of Shiga Toxigenic E. coli.69
Moreover, many studies have shown that Intimin exhibits a strong attraction for nucleolin and beta1 integrin, the eukaryotic proteins. During STEC O157:H7 infections, nucleolin and beta1 integrin serve as the most powerful receptors for Intimin.70 The TIR proteins delivered to the host cell by the LEE positive strains of STEC subsequently get inserted into the host membrane, and its extracellular region is revealed to attach Intimin. This attachment leads to the formation of multimers, resulting in an outstanding Tir clustering terminating in a signal which generates actin polymerization that operates actin pedestal formation.71 The adhesion induces the above-mentioned A/E lesions. This lesion is controlled by the Locus of Enterocyte Effacement (LEE), a large Pathogenicity Island.72 The products of LEE are type III secretion system, intimin receptors, intimin and some secreted proteins. Intimin is coded by the eae gene. The gene encoding intimin was perceived more often in stx1-positive strains. It is found that the eae gene also perform a great job in pathogenesis of Enteropathogenic E. coli (EPEC) in human beings.73 The exhibition of tropism by O157:H7 serotypes were reported in several studies in the terminal rectum of the bovines, especially the 3 to 5 centimeter close to RAJ (Recto Anal Junction). From these findings, they put forwarded a hypothesis that super shedders, a subset of cattle, are capable of shedding the shigatoxigenic E.coli serotype, the O157:H7 strains at a rate of about 104 cfu/gm of fecal matter and the factor responsible for this super shedding is colonization at RAJ.74 Many studies also reported the influence of many effectors that are non-LEE encoded such as EspJ, NleB, NleE, NleF, NleH, etc. are found in EHEC survival and colonization.8 During EPEC and EHEC infection, it was found that NleE plays the main role in the natural inborn immune response modulation through inducing some decrease in the expression as well as the production of the cytokine IL-8, NleH functions as the translocated opponent of the pro-apoptotic outcomes produced by enteropathogenic/enterohemorrhagic types, thereby promoting cell survival and sustained colonization of EHEC while NleB is found to interfere with the inflammatory signaling pathways.75
Autoagglutinating adhesin
Studies conducted on a LEE negative STEC strain, 0113:H21, which is responsible for HUS outbreaks, led to the isolation of a gene from its mega plasmid, that is responsible for encoding an adhesin which is self-agglutinating named Saa (STEC auto-agglutinating adhesin), an additional virulence factor.76,77 The exhibition of significant variation in size of Saa proteins coded by saa genes is also interesting, and the reason may be the deletion of direct repeat units, either one or two, length ranging from 460 to 53 amino acids. More evidences are there for supporting the adhesin function of saa. Primarily, a significant reduction in adherence property was observed in the plasmid cured and Saa negative mutant derivatives. The adhesion is not completely abolished, proving the role of some other factors contributing to other adhesion mechanisms. Studies also revealed that the anti-Saa antibody was capable of inhibiting the adherence property; for this reason, this self-agglutinating adhesin must be more focused and studied more as a strong antigen that can be employed for vaccine production for STEC strains that are LEE negative. Ultimately, the purified exogenous protein is expected to compete to bind on the specific sites on the surface of the epithelial cell with the endogenous adhesins. The presence of coiled-coil domains in Saa is also credible as the external proteins would be able to interact directly with Saa, which is located on the surface, thereby enhancing their interaction with other bacterial cells or epithelial cells.78
Enterohemolysins
Enterohemolysins are plasmid-encoded additional virulence-associated markers.79 This protein toxin is another virulence factor present in O157:H7 STEC strains. Enterohemolysin can damage the membrane of RBCs and can be employed as an alternate mechanism for the detection of STEC E. coli.59 Mainly four classes of hemolysins are recognized namely Alpha-hemolysin, (hlyA), plasmid-carried enterohemolysin (ehxA), phage-associated enterohemolysin (e-hlyA) and silent hemolysin (she A).59 Enterohemolysin shows the property of hemolysis on the washed sheep blood agar, which can be considered as a phenotypic indicator of the shigatoxigenic strains of E. coli.80 It is also noticeable that the plasmid carried enterohemolysin exhibits a very close association with the strains responsible for diarrhea and HUS.15,81 Due to above mentioned reasons exhA is considered as the “epidemiological marker” that can be utilized for the characterization of EHEC strains.82,83 The nucleotide sequence of ehxA contains approximately 3000 base pairs, and it is located in the ehx cluster. The ehx cluster is seen in pO157 plasmid and consists of four genes ehxC, ehxA, ehxB, and ehxD.84
Catalase-peroxidase (kat P)
Studies revealed that a plasmid-derived catalase-peroxidase was also present in the O157:H7 strain of EHEC, in addition to the chromosome encoded catalase-peroxidases.74 The encoding gene was isolated from a 9-7 kb DNA fragment derived from pO157( large plasmid of O157:H7 strain EDL 933 of EHEC).85 An Open Reading Frame of about 2208 base pairs and a polypeptide with 736 amino acid residues having a molecular mass of 81.8 kDa was found in nucleotide sequencing. This supposed protein showed very high similarity to the catalase-peroxidase family of bacteria. Moreover, the detailed examination of the peptide sequence of the protein divulged the existence of type I and II peroxidase motifs. The by-products of oxygen metabolism, Reactive oxygen molecules can damage the bacteria in several ways. Recently, it has been strongly considered that the enzymes, catalase and superoxide dismutase produced by the bacterial cells are capable of protecting these bacteria from oxidative damage brought about by the reactive oxygen molecules, the products of phagocytes and host cells.86
Extracellular serine protease
In several Gram-negative bacteria, including E. coli, a growing family of serine protease was detected. They are secreted externally by the auto transporter pathway. These enzymes come under ‘SPATEs’ family (Serine Protease Autotransporter of Enterobacteriaceae family). This plasmid-encoded protein (EspP), was first reported in 1997 as a secreted protein and was associated with a plasmid (pO157) of E. coli O157:H7.87 Currently, five subtypes of EspP recognized in Shiga toxin-producing E. coli, namely α, β, γ, δ, and ε.88-90 EspPα is connected with some extremely pathogenic serotypes such as O26, O111, O145, and O157, therefore, considered an additional virulence factor.90
EspP consists of an N-terminal signal peptide with 55 amino acid residues, one secreted passenger domain having 56-1023 amino acids and the C-terminal β-domain, which has a length of 277 amino acids, also known as translocator. The passenger domain bears a linker-region of about 30 amino acids at the C-terminal region. This Linker region connects the β-domain and the passenger domain, which is also necessary for the folding of the β-domain and its stability along with the auto chaperone motif seen at the Passenger domain C-terminal (Figure 6).91 The beta-domain directs the protein to the outer membrane of bacteria and aids the translocation of the N terminal of the Passenger domain across the membrane. Then autoproteolytic separation of the Passenger domain from the beta- domain occurs, resulting in the release of mature protein of 104 kDa size, exhibiting serine protease activity in the extracellular environment Within the N terminal domain, the proteolytic ability of the protein is confined.91 The passenger domain also has a β-helical stalk domain which is an expanded right-handed parallel one whose functional role is still unclear.
Figure 6. Structural Organization of EspP. EspP consists of an N-terminal signal peptide with 55 amino acid residues, one secreted passenger domain having 56-1023 amino acids and the C-terminal β-domain, which has a length of 277 amino acids, also known as translocator. The passenger domain bears a linker-region of about 30 amino acids at the C-terminal region. This Linker region connects the β-domain and the passenger domain, which is also necessary for the folding of the β-domain and its stability along with the auto chaperone motif seen at the C terminal of the passenger domain.91
Zinc metalloprotease (stcE)
Another important putative virulence factor is zinc metalloprotease (StcE) produced by the O157:H7 strain of EHEC92, released by the Type-II secretion system that is coded on pO157 virulence plasmid, which is offering great support for mammalian colonization by proteolytically exposing the cell surface of the host leading to close adherence of the organism to the host cell.74,93 The present representation for the action of StcE states that primarily, StcE permits the movement of the bacterium along the oral cavity by splitting the mucin-type glycoprotein present in saliva utilizing its metalloprotease-mediated mucinase activity that is accountable for the aggregation of bacteria. By splitting glycoprotein 340 as well as mucin existing in saliva, it can break down and lessen the thickness of the mucus layer.94 Likewise, StcE cleaves the glycoprotein which protect the epithelial surface of the intestine, thereby permitting the bacterium to create a close interaction with host’s plasma membrane, where elements of LEE mediate development of the peculiar A/E lesions. It is also found that StcE is capable of focalizing the inflammatory regulator C1 esterase inhibitor (C1-INH) to plasma membrane of the host cell, thus diminishing the complement-mediated lysis of both the bacterium and the host cell.94,95
Subtilase cytotoxin
SubAB, a member of the AB5 cytotoxin family was detected in a serotype O113:H21 of STEC strains lacking the Locus of Enterocyte Effacement in association with the outburst of Hemolytic Uremic Syndrome in South Australia96 Affected patients showed unusual symptoms such as neurological disorders. SubABI and Sub ABII are the two forms of subtilase cytotoxin whose coding genes were positioned on either chromosome or plasmids, respectively. The first reported toxin Sub ABI was encrypted on the virulence plasmid, pO113, present in the O113:H21 strain.97 Additionally, SubAB2 was grouped into three subsets, SubAB2-1, I, and III. Of these three subtypes, subtype-I is encoded on SE-PAI (pathogenicity island).98 The subtype-II is encoded in a locus on the efflux protein of the outer membrane.95 The last subtype was seen closely associated with the gene that is encoding one protein whose function is not known yet.99 Studies revealed that the collaborative action of both Stx2 and SubAB is capable of causing more severe renal impairment and the occurrence of typical symptoms of HUS such as partial or total destruction of kidneys, erythrocyte variation, the rise of free hemoglobin etc. in humans.96
Acid-resistance mechanisms
Acid-resistance mechanisms are both enzyme and chaperone based. Mainly three major enzyme-based acid-resistance mechanisms are there for O157 STEC strains which protect them from acidic pH 2.0 to 2.5, enabling the strains of E. coli in withstanding extremely acidic environment experienced while travelling via the host stomach, and also to long-term exposure to even the mild acidic conditions of the host intestine. Acid resistance mechanism 1 is induced by sigma factor, the stationary phase alternative and cAMP receptor protein (CRP), the global regulatory protein. Acid resistance mechanism-2 is the glutamate decarboxylase system needs glutamate, genes encrypting glutamate decarboxylase (gad A, gad B, and gad C gene), as well as g-aminobutyric acid antiporter, in order to safeguard the cells from extremely acidic condition. Acid resistance mechanism-3 is the arginine decarboxylase system, which needs amino acid arginine, adiA (arginine decarboxylase gene), adiC (arginine/ agmatine antiporter gene), and one regulator cysB. Lysine and ornithine decarboxylase systems are other acid resistance mechanisms seen in E. coli whose role is not yet revealed.100,101 Other important genes associated with acid resistance are seen on AFI (Acid Fitness Island). AFI has a size of 15 kb, located on the chromosome of the bacterium. It is controlled and repressed by Rpos, stationary phase sS factor and histone-like nucleoid-structure protein, respectively.8,102 Studies revealed that in a few non-O157 STEC strains, to combat acidic conditions. HdeA and HdeB are the main chaperones.103 STEC strains utilize a Hydrogenase-3-based acid resistance mechanism to combat acidic conditions anaerobically. Studies also found that in STEC O157:H7, the DNA-binding proteins occurring in starved cells (Dps) play well in acid tolerance. In addition to this, Dps has many other functions such as iron sequestration reduction of oxidative stress-mediated by iron etc.104 Acid resistance will surely enhance the survival of these strains in food items such as sausages, dairy products, fruit juices etc.
Iron Acquisition
Studies revealed that, like other bacteria, STEC also possess some mechanisms for acquiring iron according to their need from the iron reserves of the host.105 Examination of several STEC strains showed that all of the strains carried an iron chelator which can bind and transport into the cell. Enterobactin/enterochelin, the catechol siderophore is the Fe-chelator in O157 STEC strains. The aerobactin, hydroxamate siderophore, is not recognized in STEC O157 strain. The presence of aerobactin and yersiniabactin was noted in a few non-O157/H— strains.106-108
Shigatoxigenic E. coli are highly virulent pathogens causing serious complications and severe diseases in humans all over the world. Their pathogenicity is a multifactorial process, and besides the well-known Shiga toxins, EHEC expresses various virulence factors. From various studies, it is very clear that there is a complex interaction occurring in biofilm formation. Moreover a comprehensive network playing an important role in virulence can be seen in STEC biofilm formation. The specific cell surface determinants of the bacterial strains also exhibit pivotal performance in the developmental steps of the biofilm. In addition to this, biofilm formation is also influenced greatly by environmental conditions and many other challenges. Due to the development of escalated resistance exhibited by the biofilm community to multiple drugs, disinfectants, sanitizers, etc., the introduction of highly improved ideas and strategies for regulating biofilm is inevitable. Another point to be considered very seriously is the initiative to design much more efficient antibiotics and disinfecting agents so that we can remove the biofilms more effectively. Another strategy that can be adopted is the use of enzymes as a supplement to disinfecting solutions. More and more research works must be done in identifying new effective agents through the detailed analysis of the biofilm structure, composition and its properties.
ACKNOWLEDGMENTS
None.
CONFLICT OF INTEREST
The authors declare that there is no conflict of interest.
AUTHORS’ CONTRIBUTION
Both authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
FUNDING
None.
DATA AVAILABILITY
All datasets generated or analyzed during this study are included in the manuscript.
ETHICS STATEMENT
Not applicable.
- Kaper JB, O’brien AD. Overview and Historical Perspectives. Microbiol Spectr. 2014;2(2).
Crossref - Thierry SIL, Gannon JE, Jaufeerally-Fakim Y, Santchurn SJ. Shiga-toxigenic Escherichia coli from animal food sources in Mauritius: Prevalence, serogroup diversity and virulence profiles. Int J Food Microbiol. 2020;324:108589.
Crossref - Dehkordi FS, Yazdani F, Mozafari J, Valizadeh Y. Virulence factors, serogroups and antimicrobial resistance properties of Escherichia coli strains in fermented dairy products. BMC Research Notes. 2014;7(1):217.
Crossref - Momtaz H, Farzan R, Rahimi E, Safarpoor Dehkordi F, Souod N. Molecular characterization of Shiga toxin-producing Escherichia coli isolated from ruminant and donkey raw milk samples and traditional dairy products in Iran. ScientificWorldJournal. 2012;231342.
Crossref - Momtaz H, Dehkordi FS, Rahimi E, Ezadi H, Arab R. Incidence of Shiga toxin-producing Escherichia coli serogroups in ruminant’s meat. Meat Sci. 2013;95(2):381-388.
Crossref - Naylor SW, Low JC, Besser TE, et al. Lymphoid follicle-dense mucosa at the terminal rectum is the principal site of colonization of enterohemorrhagic Escherichia coli O157:H7 in the bovine host. Infect Immun. 2003;71(3):1505-1512.
Crossref - Pruimboom-Brees IM, Morgan TW, Ackermann MR, et al. Cattle lack vascular receptors for Escherichia coli O157:H7 Shiga toxins. Proc Natl Acad Sci U S A. 2000;97(19):10325-10329.
Crossref - Sperandio V, Nguyen Y. Enterohemorrhagic E. coli (EHEC) pathogenesis. Front Cell Infect Microbiol. 2012;2:90.
Crossref - Gyles CL. Shiga toxin-producing Escherichia coli: an overview. J Anim Sci. 2007;85(13 Suppl):E45-62.
Crossref - Newell DG, La Ragione RM. Enterohaemorrhagic and other Shiga toxin-producing Escherichia coli (STEC): Where are we now regarding diagnostics and control strategies? Transbound Emerg Dis. 2018;65(Suppl 1):49-71.
Crossref - Ferens WA, Hovde CJ. Escherichia coli O157:H7: animal reservoir and sources of human infection. Foodborne Pathog Dis. 2011;8(4):465-487.
Crossref - Kim JS, Lee MS, Kim JH. Recent Updates on Outbreaks of Shiga Toxin-Producing Escherichia coli and Its Potential Reservoirs. Front Cell Infect Microbiol. 2020;10:273.
Crossref - Blanco Crivelli X, Rumi M, Carfagnini J, Degregorio O, Bentancor A. Synanthropic rodents as possible reservoirs of shigatoxigenic Escherichia coli strains. Front Cell Infect Microbiol. 2012;2:134.
Crossref - Baker CA, Rubinelli PM, Park SH, Carbonero F, Ricke SC. Shiga toxin-producing Escherichia coli in food: Incidence, ecology, and detection strategies. Food Control. 2016;59:407-419.
Crossref - Joseph J, Kalyanikutty S. Occurrence of multiple drug-resistant Shiga toxigenic Escherichia coli in raw milk samples collected from retail outlets in South India. J Food Sci Technol. 2022;59(6):2150-2159.
Crossref - Hwang SB, Chelliah R, Kang JE, et al. Role of Recent Therapeutic Applications and the Infection Strategies of Shiga Toxin-Producing Escherichia coli. Front Cell Infect Microbiol. 2021;11:614963.
Crossref - Onyeka LO, Adesiyun AA, Keddy KH, Madoroba E, Manqele A, Thompson PN. Shiga Toxin-Producing Escherichia coli Contamination of Raw Beef and Beef-Based Ready-to-Eat Products at Retail Outlets in Pretoria, South Africa. J Food Prot. 2020;83(3):476-484.
Crossref - Giaouris E, Heir E, Desvaux M, et al. Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front Microbiol. 2015;6:841.
Crossref - Flemming HC, Wingender J, Szewzyk U, Steinberg P, Rice SA, Kjelleberg S. Biofilms: an emergent form of bacterial life. Nat Rev Microbiol. 2016;14(9):563-575.
Crossref - Benyoussef W, Deforet M, Monmeyran A, Henry N. Flagellar Motility During E. coli Biofilm Formation Provides a Competitive Disadvantage Which Recedes in the Presence of Co-Colonizers. Front Cell Infect Microbiol. 2022;12:896898.
Crossref - Vogeleer P, Tremblay YDN, Jubelin G, Jacques M, Harel J. Biofilm-Forming Abilities of Shiga Toxin-Producing Escherichia coli Isolates Associated with Human Infections. Appl Environ Microbiol. 2016;82(5):1448-1458.
Crossref - Taghadosi R, Shakibaie MR, Masoumi S. Biochemical detection of N-Acyl homoserine lactone from biofilm-forming uropathogenic Escherichia coli isolated from urinary tract infection samples. Rep Biochem Mol Biol. 2015;3(2):56-61.
- Taghadosi R, Shakibaie MR, Ghanbarpour R, Hosseini-Nave H. Role of antigen-43 on biofilm formation and horizontal antibiotic resistance gene transfer in non-O157 Shiga toxin producing Escherichia coli strains. Iran J Microbiol. 2017;9(2):89-96.
- Barrett TJ, Kaper JB, Jerse AE, Wachsmuth IK. Virulence factors in Shiga-like toxin-producing Escherichia coli isolated from humans and cattle. J Infect Dis. 1992;165(5):979-980.
Crossref - Pinto G, Sampaio M, Dias O, Almeida C, Azeredo J, Oliveira H. Insights into the genome architecture and evolution of Shiga toxin encoding bacteriophages of Escherichia coli. BMC Genomics. 2021;22(1):366.
Crossref - Gati NS, Middendorf-Bauchart B, Bletz S, Dobrindt U, Mellmann A. Origin and Evolution of Hybrid Shiga Toxin-Producing and Uropathogenic Escherichia coli Strains of Sequence Type 141. J Clin Microbiol. 2019;58(1):e01309-19.
Crossref - Hiroi M, Takahashi N, Harada T, et al. Serotype, Shiga toxin (Stx) type, and antimicrobial resistance of Stx-producing Escherichia coli isolated from humans in Shizuoka Prefecture, Japan (2003-2007). Jpn J Infect Dis. 2012;65(3):198-202.
- Pan Y, Hu B, Bai X, et al. Antimicrobial Resistance of Non-O157 Shiga Toxin-Producing Escherichia coli Isolated from Humans and Domestic Animals. Antibiotics. 2021;10(1):74.
Crossref - Bai L, Hurley D, Li J, et al. Characterisation of multidrug-resistant Shiga toxin-producing Escherichia coli cultured from pigs in China: co-occurrence of extended-spectrum β-lactamase- and mcr-1-encoding genes on plasmids. Int J Antimicrob Agents. 2016;48(4):445-448.
Crossref - Iweriebor BC, Iwu CJ, Obi LC, Nwodo UU, Okoh AI. Multiple antibiotic resistances among Shiga toxin producing Escherichia coli O157 in feces of dairy cattle farms in Eastern Cape of South Africa. BMC Microbiol. 2015;15:213.
Crossref - Skandalis N, Maeusli M, Papafotis D, et al. Environmental Spread of Antibiotic Resistance. Antibiotics. 2021;10(6):640.
Crossref - Larsson DGJ, Flach CF. Antibiotic resistance in the environment. Nat Rev Microbiol. 2022;20(5):257-269.
Crossref - Al-Marri T, Al-Marri A, Al-Zanbaqi R, Ajmi AA, Fayez M. Multidrug resistance, biofilm formation, and virulence genes of Escherichia coli from backyard poultry farms. Vet World. 2021;14(11):2869-2877.
Crossref - Rodis N, Tsapadikou VK, Potsios C, Xaplanteri P. Resistance Mechanisms in Bacterial Biofilm Formations: A Review 2020:30.
Crossref - Khan J, Tarar SM, Gul I, Nawaz U, Arshad M. Challenges of antibiotic resistance biofilms and potential combating strategies: a review. 3 Biotech. 2021;11(4):169.
Crossref - Vasudevan S, Joseph HA, Swamy SS, Solomon AP. Antibiotic Resistance in Biofilms. In: Rathinam NK, Sani RK, eds. ACS Symposium Series. American Chemical Society. 2019;1323:205-224.
Crossref - Gebreyohannes G, Nyerere A, Bii C, Sbhatu DB. Challenges of intervention, treatment, and antibiotic resistance of biofilm-forming microorganisms. Heliyon. 2019;5(8):e02192.
Crossref - Jo J, Price-Whelan A, Dietrich LEP. Gradients and consequences of heterogeneity in biofilms. Nat Rev Microbiol. 2022;20(10):593-607.
Crossref - Khalil RKS, Skinner C, Patfield S, He X. Phage-mediated Shiga toxin (Stx) horizontal gene transfer and expression in non-Shiga toxigenic Enterobacter and Escherichia coli strains. Pathog Dis. 2016;74(5):ftw037.
Crossref - Schroeder M, Brooks BD, Brooks AE. The Complex Relationship between Virulence and Antibiotic Resistance. Genes. 2017;8(1):39.
Crossref - Foster TJ. Antibiotic resistance in Staphylococcus aureus. Current status and future prospects. FEMS Microbiol Rev. 2017;41(3):430-449.
Crossref - Colello R, Krüger A, Conza J, et al. Antimicrobial Resistance in Class 1 Integron-Positive Shiga Toxin-Producing Escherichia coli Isolated from Cattle, Pigs, Food and Farm Environment. Microorganisms. 2018;6(4):99.
Crossref - Racewicz P, Majewski M, Madeja ZE, Lukomska A, Kubiak M. Role of integrons in the proliferation of multiple drug resistance in selected bacteria occurring in poultry production. Br Poult Sci. 2020;61(2):122-131.
Crossref - Van Meervenne E, Boon N, Verstraete K, et al. Integron characterization and typing of Shiga toxin-producing Escherichia coli isolates in Belgium. J Med Microbiol. 2013;62(5):712-719.
Crossref - Domingues S, da Silva GJ, Nielsen KM. Integrons: Vehicles and pathways for horizontal dissemination in bacteria. Mob Genet Elements. 2012;2(5):211-223.
Crossref - Hossain T, Deter HS, Peters EJ, Butzin NC. Antibiotic tolerance, persistence, and resistance of the evolved minimal cell, Mycoplasma mycoides JCVI-Syn3B. iScience. 2021;24(5):102391.
Crossref - Menge C. The Role of Escherichia coli Shiga Toxins in STEC Colonization of Cattle. Toxins. 2020;12(9):607.
Crossref - Silva CJ, Brandon DL, Skinner CB, He X. Structure of Shiga Toxins and Other AB5 Toxins. In: Silva CJ, Brandon DL, Skinner CB, He X, eds. Shiga Toxins: A Review of Structure, Mechanism, and Detection. Food Microbiology and Food Safety. 2017:21-45.
Crossref - Lee KS, Jeong YJ, Lee MS. Escherichia coli Shiga Toxins and Gut Microbiota Interactions. Toxins. 2021;13(6):416.
Crossref - Scheutz F, Teel LD, Beutin L, et al. Multicenter evaluation of a sequence-based protocol for subtyping Shiga toxins and standardizing Stx nomenclature. J Clin Microbiol. 2012;50(9):2951-2963.
Crossref - Lee MS, Koo S, Jeong DG, Tesh VL. Shiga Toxins as Multi-Functional Proteins: Induction of Host Cellular Stress Responses, Role in Pathogenesis and Therapeutic Applications. Toxins. 2016;8(3):77.
Crossref - Melton-Celsa AR. Shiga Toxin (Stx) Classification, Structure, and Function. Microbiol Spectr. 2014;2(4):EHEC-0024-2013.
Crossref - Walsh MJ, Dodd JE, Hautbergue GM. Ribosome-inactivating proteins. Virulence. 2013;4(8):774-784.
Crossref - Zhou Y, Li XP, Kahn JN, Tumer NE. Functional Assays for Measuring the Catalytic Activity of Ribosome Inactivating Proteins. Toxins. 2018;10(6):240.
Crossref - Melton-Celsa A, Mohawk K, Teel L, O’Brien A. Pathogenesis of Shiga-toxin producing escherichia coli. Curr Top Microbiol Immunol. 2012;357:67-103.
Crossref - Farfan MJ, Torres AG. Molecular Mechanisms That Mediate Colonization of Shiga Toxin-Producing Escherichia coli Strains. Infect Immun. 2012;80(3):903-913.
Crossref - Haiko J, Westerlund-Wikström B. The Role of the Bacterial Flagellum in Adhesion and Virulence. Biology. 2013;2(4):1242-1267.
Crossref - Edwards RA, Puente JL. Fimbrial expression in enteric bacteria: a critical step in intestinal pathogenesis. Trends Microbiol. 1998;6(7):282-287.
Crossref - Lorenz SC, Son I, Maounounen-Laasri A, Lin A, Fischer M, Kase JA. Prevalence of Hemolysin Genes and Comparison of ehxA Subtype Patterns in Shiga Toxin-Producing Escherichia coli (STEC) and Non-STEC Strains from Clinical, Food, and Animal Sources. Appl Environ Microbiol. 2013;79(20):6301-6311.
Crossref - Clarke KR, Hor L, Pilapitiya A, Luirink J, Paxman JJ, Heras B. Phylogenetic Classification and Functional Review of Autotransporters. Front Immunol. 2022;13:1272.
Crossref - Vo JL, Martinez Ortiz GC, Subedi P, et al. Autotransporter Adhesins in Escherichia coli Pathogenesis. Proteomics. 2017;17(23-24):1600431.
Crossref - Carter MQ, Louie JW, Feng D, Zhong W, Brandl MT. Curli fimbriae are conditionally required in Escherichia coli O157:H7 for initial attachment and biofilm formation. Food Microbiol. 2016;57:81-89.
Crossref - Loof TG, Deicke C, Medina E. The role of coagulation/fibrinolysis during Streptococcus pyogenes infection. Front Cell Infect Microbiol. 2014;4:128.
Crossref - Uhlich GA, Chen CY, Cottrell BJ, Nguyen LH. Growth media and temperature effects on biofilm formation by serotype O157:H7 and non-O157 Shiga toxin-producing Escherichia coli. FEMS Microbiol Lett. 2014;354(2):133-141.
Crossref - Ageorges V, Monteiro R, Leroy S, et al. Molecular determinants of surface colonisation in diarrhoeagenic Escherichia coli (DEC): from bacterial adhesion to biofilm formation. FEMS Microbiol Rev. 2020;44(3):314-350.
Crossref - Guo XP, Sun YC. New Insights into the Non-orthodox Two Component Rcs Phosphorelay System. Front Microbiol. 2017;8.
Crossref - Alalam H, Graf FE, Palm M, et al. A High-Throughput Method for Screening for Genes Controlling Bacterial Conjugation of Antibiotic Resistance. mSystems. 2020;5(6):e01226-20.
Crossref - Virolle C, Goldlust K, Djermoun S, Bigot S, Lesterlin C. Plasmid Transfer by Conjugation in Gram-Negative Bacteria: From the Cellular to the Community Level. Genes (Basel). 2020;11(11):1239.
Crossref - Hartland E, Leong J. Enteropathogenic and enterohemorrhagic E. coli: ecology, pathogenesis, and evolution. Front Cell Infect Microbiol. 2013;3.
Crossref - McWilliams BD, Torres AG. Enterohemorrhagic Escherichia coli Adhesins. Microbiol Spectr. 2014;2(3).
Crossref - Nascimento HH, Silva LE, Souza RT, Silva NP, Scaletsky IC. Phenotypic and genotypic characteristics associated with biofilm formation in clinical isolates of atypical enteropathogenic Escherichia coli (aEPEC) strains. BMC Microbiol. 2014;14(1):184.
Crossref - Franzin FM, Sircili MP. Locus of Enterocyte Effacement: A Pathogenicity Island Involved in the Virulence of Enteropathogenic and Enterohemorragic Escherichia coli Subjected to a Complex Network of Gene Regulation. BioMed Res Int. 2015;e534738.
Crossref - Yang X, Sun H, Fan R, et al. Genetic diversity of the intimin gene (eae) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci Rep. 2020;10(1):3275.
Crossref - Etcheverria AI, Padola NL. Shiga toxin-producing Escherichia coli. Virulence. 2013;4(5):366-372.
Crossref - Pollock GL, Oates CVL, Giogha C, et al. Distinct Roles of the Antiapoptotic Effectors NleB and NleF from Enteropathogenic Escherichia coli. Infect Immun. 2017;85(4):e01071-16.
Crossref - Mohseni M, Djawadi B, Khazaei N. Escherichia Coli O157:H7 and Its Effect on Human Health. IntechOpen. 2022.
Crossref - Sokolovic M, Simpraga B, Amsel-Zelenika T, Berendika M, Krstulovic F. Prevalence and Characterization of Shiga Toxin Producing Escherichia coli Isolated from Animal Feed in Croatia. Microorganisms. 2022;10(9):1839.
Crossref - Paton AW, Srimanote P, Woodrow MC, Paton JC. Characterization of Saa, a novel autoagglutinating adhesin produced by locus of enterocyte effacement-negative Shiga-toxigenic Escherichia coli strains that are virulent for humans. Infect Immun. 2001;69(11):6999-7009.
Crossref - Tayh G, Boubaker SM, Khedher RB, et al. Prevalence, virulence genes, and antimicrobial profiles of Escherichia coli O157:H7 isolated from healthy cattle in Tunisia. J Infect Dev Ctries. 2022;16(8):1308-1316.
Crossref - Hua Y, Zhang J, Jernberg C, et al. Molecular Characterization of the Enterohemolysin Gene (ehxA) in Clinical Shiga Toxin-Producing Escherichia coli Isolates. Toxins. 2021;13(1):71.
Crossref - Ethelberg S, Olsen KEP, Scheutz F, et al. Virulence factors for hemolytic uremic syndrome, Denmark. Emerg Infect Dis. 2004;10(5):842-847.
Crossref - Beutin L, Montenegro MA, Orskov I, et al. Close association of verotoxin (Shiga-like toxin) production with enterohemolysin production in strains of Escherichia coli. J Clin Microbiol. 1989;27(11):2559-2564.
Crossref - Haque M, Bosilevac J, Chaves B. A review of Shiga-toxin producing Escherichia coli (STEC) contamination in the raw pork production chain. Int J Food Microbiol. 2022;377:109832.
Crossref - Fu S, Bai X, Fan R, Sun H, Xu Y, Xiong Y. Genetic diversity of the enterohaemolysin gene (ehxA) in non-O157 Shiga toxin-producing Escherichia coli strains in China. Sci Rep. 2018;8(1):4233.
Crossref - Latif H, Li HJ, Charusanti P, Palsson BO, Aziz RK. A Gapless, Unambiguous Genome Sequence of the Enterohemorrhagic Escherichia coli O157:H7 Strain EDL933. Genome Announcements. 2014;2(4):e00821-14.
Crossref - Cheng JH, Lv X, Pan Y, Sun DW. Foodborne bacterial stress responses to exogenous reactive oxygen species (ROS) induced by cold plasma treatments. Trends Food Sci Technol. 2020;103:239-247.
Crossref - Brunder W, Schmidt H, Karch H. EspP, a novel extracellular serine protease of enterohaemorrhagic Escherichia coli O157:H7 cleaves human coagulation factor V. Mol Microbiol. 1997;24(4):767-778.
Crossref - Bielaszewska M, Stoewe F, Fruth A, et al. Shiga Toxin, Cytolethal Distending Toxin, and Hemolysin Repertoires in Clinical Escherichia coli O91 Isolates. J Clin Microbiol. 2009;47(7):2061-2066.
Crossref - Brockmeyer J, Bielaszewska M, Fruth A, et al. Subtypes of the Plasmid-Encoded Serine Protease EspP in Shiga Toxin-Producing Escherichia coli: Distribution, Secretion, and Proteolytic Activity. Appl Environ Microbiol. 2007;73(20):6351-6359.
Crossref - Pokharel P, Habouria H, Bessaiah H, Dozois CM. Serine Protease Autotransporters of the Enterobacteriaceae (SPATEs): Out and About and Chopping It Up. Microorganisms. 2019;7(12):594.
Crossref - Weiss A, Brockmeyer J. Prevalence, Biogenesis, and Functionality of the Serine Protease Autotransporter EspP. Toxins. 2013;5(1):25-48.
Crossref - Kobayashi N, Lee K ichi, Yamazaki A, et al. Virulence Gene Profiles and Population Genetic Analysis for Exploration of Pathogenic Serogroups of Shiga Toxin-Producing Escherichia coli. J Clin Microbiol. 2013;51(12):4022-4028.
Crossref - Lathem WW, Bergsbaken T, Welch RA. Potentiation of C1 esterase inhibitor by StcE, a metalloprotease secreted by Escherichia coli O157:H7. J Exp Med. 2004;199(8):1077-1087.
Crossref - Yu ACY, Worrall LJ, Strynadka NCJ. Structural Insight into the Bacterial Mucinase StcE Essential to Adhesion and Immune Evasion during Enterohemorrhagic E. coli Infection. Structure. 2012;20(4):707-717.
Crossref - Funk J, Stoeber H, Hauser E, Schmidt H. Molecular analysis of subtilase cytotoxin genes of food-borne Shiga toxin-producing Escherichia coli reveals a new allelic subAB variant. BMC Microbiol. 2013;13(1):230.
Crossref - Seyahian EA, Oltra G, Ochoa F, et al. Systemic effects of Subtilase cytotoxin produced by Escherichia coli O113:H21. Toxicon. 2017;127:49-55.
Crossref - Paton AW, Woodrow MC, Doyle RM, Lanser JA, Paton JC. Molecular characterization of a Shiga toxigenic Escherichia coli O113:H21 strain lacking eae responsible for a cluster of cases of hemolytic-uremic syndrome. J Clin Microbiol. 1999;37(10):3357-3361.
Crossref - Michelacci V, Tozzoli R, Caprioli A, et al. A new pathogenicity island carrying an allelic variant of the Subtilase cytotoxin is common among Shiga toxin producing Escherichia coli of human and ovine origin. Clin Microbiol Infect. 2013;19(3):E149-156.
Crossref - Tasara T, Fierz L, Klumpp J, Schmidt H, Stephan R. Draft Genome Sequences of Five Shiga Toxin-Producing Escherichia coli Isolates Harboring the New and Recently Described Subtilase Cytotoxin Allelic Variant subAB2-3. Genome Announc. 2017;5(8):e01582-16.
Crossref - Foster JW. Escherichia coli acid resistance: tales of an amateur acidophile. Nat Rev Microbiol. 2004;2(11):898-907.
Crossref - Kanjee U, Houry WA. Mechanisms of Acid Resistance in Escherichia coli. Ann Rev Microbiol. 2013;67(1):65-81.
Crossref - Tramonti A, De Canio M, De Biase D. GadX/GadW-dependent regulation of the Escherichia coli acid fitness island: transcriptional control at the gadY-gadW divergent promoters and identification of four novel 42 bp GadX/GadW-specific binding sites. Mol Microbiol. 2008;70(4):965-982.
Crossref - Smith JL, Fratamico PM. Effect of stress on non-O157 Shiga toxin-producing Escherichia coli. J Food Prot. 2012;75(12):2241-2250.
Crossref - Calhoun LN, Kwon YM. Structure, function and regulation of the DNA-binding protein Dps and its role in acid and oxidative stress resistance in Escherichia coli: a review. J Appl Microbiol. 2011;110(2):375-386.
Crossref - Saha R, Saha N, Donofrio RS, Bestervelt LL. Microbial siderophores: a mini review. J Basic Microbiol. 2013;53(4):303-317.
Crossref - Barnard TJ, Dautin N, Lukacik P, Bernstein HD, Buchanan SK. Autotransporter structure reveals intra-barrel cleavage followed by conformational changes. Nat Struct Mol Biol. 2007;14(12):1214-1220.
Crossref - Dautin N, Barnard TJ, Anderson DE, Bernstein HD. Cleavage of a bacterial autotransporter by an evolutionarily convergent autocatalytic mechanism. EMBO J. 2007;26(7):1942-1952.
Crossref - Searle LJ. Population Structure and Siderophore Production in Commensal Escherichia coli. :271.
© The Author(s) 2024. Open Access. This article is distributed under the terms of the Creative Commons Attribution 4.0 International License which permits unrestricted use, sharing, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.